Core Tip: The cotton team has further mapped 3D genome maps of cotton diploid and tetraploid based on the existing 3D genome study. This study advances our understanding of chromatin organization in plants and sheds new light on the relationship between 3D genome evolution and transcriptional regulation.
(by Maojun Wang) The achievements of 3D genome researched by the cotton team of HZAU was published on Nature Plants, On January 29, 2018.The paper entitled “Evolutionary dynamics of 3D genome architecture following polyploidization in cotton”, which is an important development of the evolution of the cotton genome. And the established 3D genomic map will promote the functional genome research in cotton.
Polyploid plants are widespread in nature. The formation of polyploids significantly increases the complexity of transcriptional regulation, which is expected to be reflected in sophisticated higher-order chromatin structures. Recently, 3D genome research is a hotspot in human and animals. 3D genome studies have also been carried out in model plants, but 3D genome dynamics mediated by polyploidy events have not been reported.Cultivated tetraploid cotton (G.hirsutum and G.barbadense), originated from inter-genomic hybridization between a putative A-genome diploid (G.arboreum or G.herbaceum) and a putative D-genome diploid (G.raimondii) approximately one to two million years ago. Now, diploid and tetraploid cotton genome sequencing has been completed.
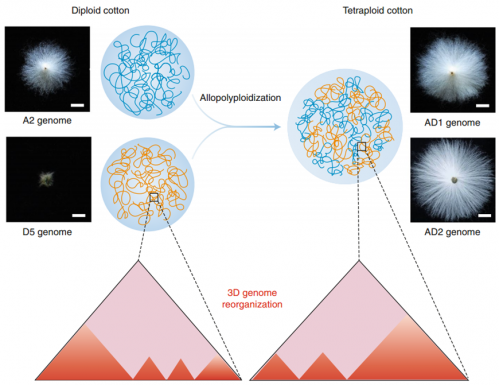
Here, we characterize 3D genome architectures for diploid and tetraploid cotton based on the existing 3D genome study(Wang et al, Nature Genetics, 2017)and find the existence of A/B compartments and topologically associated domains (TADs).By comparing each subgenome in tetraploids with its extant diploid progenitor, we find that genome allopolyploidization has contributed to the switching of A/B compartments and the reorganization of TADs in both subgenomes.We also show that the formation of TAD boundaries during polyploidization preferentially occurs in open chromatin, coinciding with the deposition of active chromatin modification.Furthermore, analysis of inter-subgenomic chromatin interactions has revealed the spatial proximity of homoeologous genes, possibly associated with their coordinated expression.This study advances our understanding of chromatin organization in plants and sheds new light on the relationship between 3D genome evolution and transcriptional regulation.The work was cooperatively achieved by the cotton team and the College of Information Science. Prof. Xianlong Zhang, Prof. Qingyong Yang and Dr. Min Lin are the corresponding author, Dr Maojun Wang is the first author.The research was supported by National Natural Science Foundation of China; the National Science Foundation for Post-doctoral Scientists of China; National Key Research and Development Program of China.